When examining the vast tapestry of genetic diversity that underpins all living organisms, a recurring question emerges: why does the concept of linkage equilibrium matter so profoundly in understanding evolutionary processes and population health? This inquiry, at its core, probes the very mechanisms by which genes, inherited together or independently, shape the adaptive potential of species. Unraveling this mystery begins with revealing the interplay between genetic loci—specifically, how their associations or dissociations influence genetic variation and the trajectory of evolution.
Deciphering Linkage Equilibrium: The Foundation of Genetic Independence
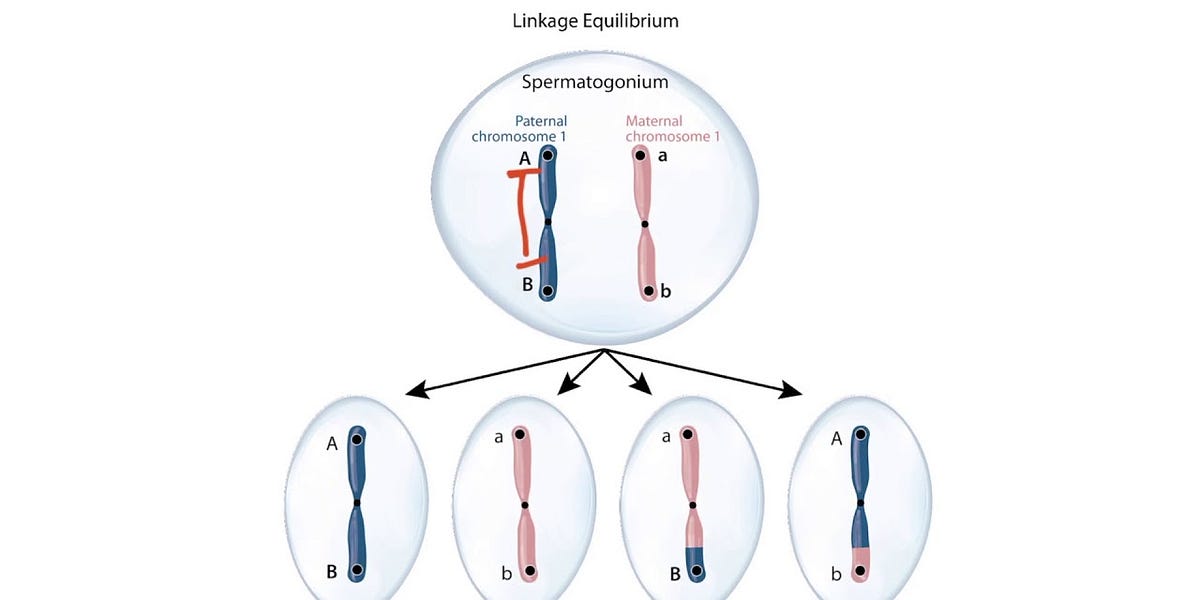
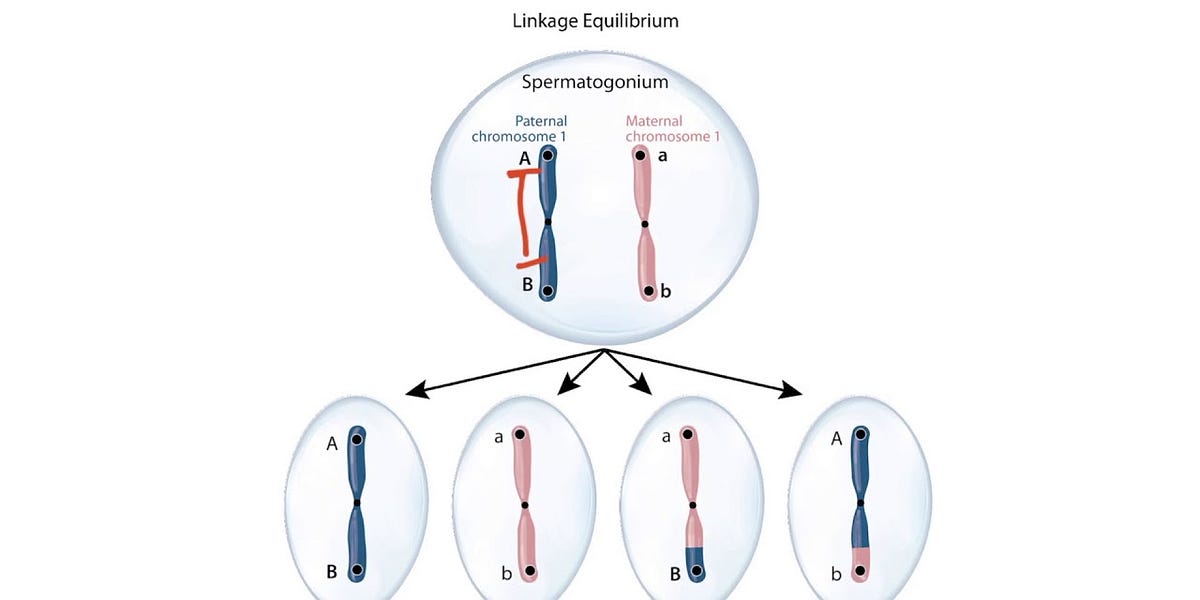
Linkage equilibrium (LE) describes a state in a population where alleles at different loci are inherited independently of each other. When a population exhibits LE, the combination of alleles at one locus bears no predictable relationship to alleles at another locus. This independence is crucial because it signifies that genetic variants can shuffle freely through recombination, fostering a rich mosaic of genotypes that enhances adaptive flexibility. Conversely, linkage disequilibrium (LD) reflects a non-random association, often a byproduct of recent admixture, selection, or genetic drift.
In-Depth: The Mechanics of Linkage Equilibrium and Disequilibrium
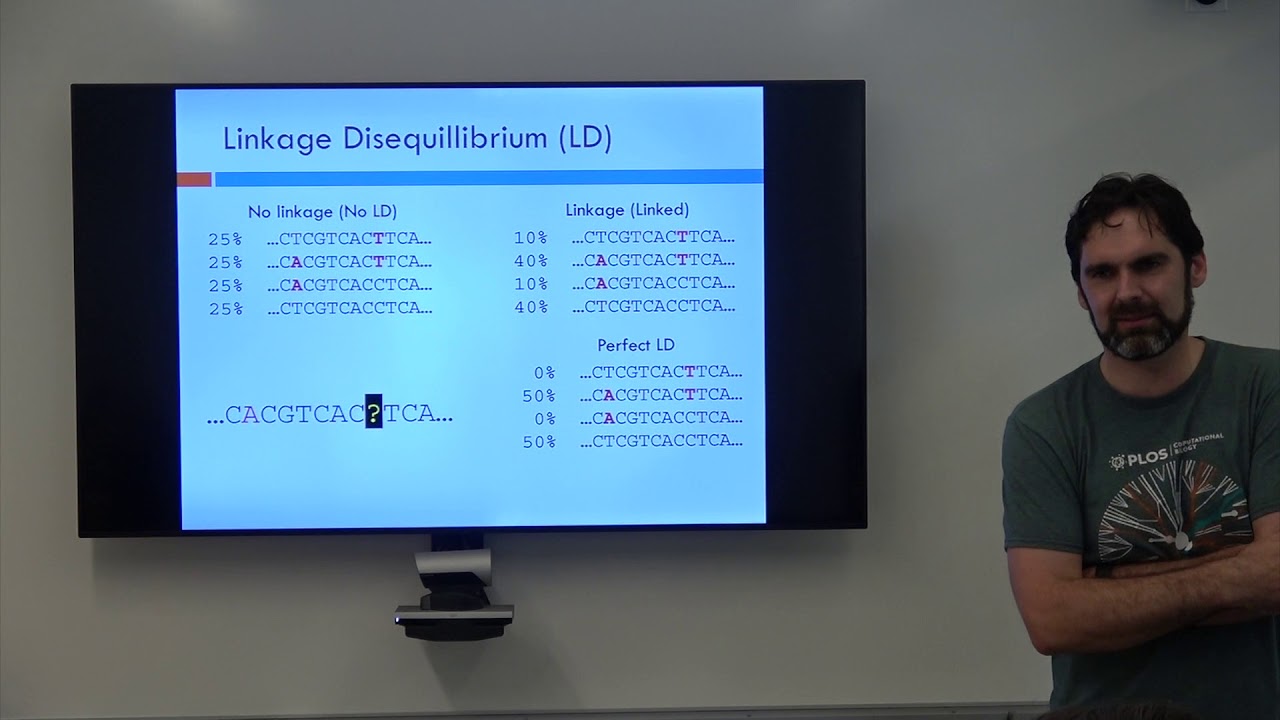
Linkage disequilibrium manifests when the observed frequency of a particular allele combination deviates from what would be expected if alleles assorted independently. For instance, in a population where alleles A/a and B/b at two loci are inherited independently, the genotype combinations like AB, Ab, aB, and ab distribute according to their respective frequencies. When LD persists, certain combinations become overrepresented, indicating a non-random association. The duration of LD depends on factors such as recombination rate, population size, and selection pressures. Understanding these dynamics is essential for grasping how populations evolve over time.
| Relevant Category | Substantive Data |
|---|---|
| Recombination Rate | The probability of crossover between two loci per generation, varies between 0 and 0.5 (50%) |
| Decay of LD | Inversely proportional to recombination rate and time; LD halves roughly every 1/(recombination rate) generations without selection or migration |
| Population Size Impact | Larger populations tend to maintain LD for shorter periods, promoting equilibrium faster due to increased recombination events |
The Evolutionary Significance of Linkage Equilibrium in Maintaining Genetic Diversity
Maintaining genetic diversity is fundamental to a species’ ability to adapt to changing environments. When linkage equilibrium prevails, alleles can undergo recombinational shuffling, producing novel genetic variants that could confer survival advantages. This process accelerates evolution and reduces the risk of genetic bottlenecks leading to extinction. In contrast, persistent linkage disequilibrium can restrict genetic variation, which may hamper adaptive responses but also harbor important signals of recent selection or structure within populations.
Implications for Population Structure and Disease Resistance
In plant and animal breeding, recognizing linkage equilibrium helps in designing effective strategies for hybridization and genetic improvement. For example, in crop populations, breaking down LD around beneficial alleles enables precise introgression and maximizes yield traits. Similarly, in human populations, the concept helps identify genomic regions under selection, such as those associated with disease resistance. It facilitates understanding how linked genes co-influence traits, offering insights into complex disease etiologies.
| Relevant Category | Substantive Data |
|---|---|
| Genetic Drift | Can generate or erode LD depending on population size and stochastic events |
| Selection | Selective sweeps can create or extend LD around advantageous alleles |
| Recombination Hotspots | Regions with high recombination rates tend to quickly attain LE |
Historical Context and the Evolution of Linkage Concepts
The foundational ideas surrounding linkage and equilibrium trace back to Gregor Mendel’s pioneering work and subsequent contributions by geneticists such as R. A. Fisher, J. B. S. Haldane, and Sewall Wright. Wright, in particular, elucidated the concept of genetic drift interacting with linkage disequilibrium, emphasizing the dynamic balance between forces maintaining or disrupting equilibrium. The advent of molecular markers like microsatellites and SNPs has allowed for practical measurement and modeling of linkage patterns across genomes, paving the way for modern population genetics and evolutionary biology.
Methodological Advances in Analyzing Linkage Dynamics
State-of-the-art tools utilize statistical methods such as the blocks of LD analysis, haplotype reconstruction, and coalescent modeling to comprehend linkage patterns. Sequencing technologies enable high-resolution mapping, revealing how recombination—the engine driving toward equilibrium—operates across complex genomes. These insights inform conservation strategies, molecular breeding, and understanding pathogen evolution, especially in rapidly changing environments.
| Relevant Category | Substantive Data |
|---|---|
| Evolutionary Models | Coalescent theory models the genealogical history of alleles, incorporating linkage and recombination |
| Genomic Technologies | Next-generation sequencing provides dense marker data to monitor linkage dynamics at the genome-wide level |
| Statistical Tools | Measures like D', r², and haplotype diversity quantify linkage disequilibrium strength |
Practical Applications and Future Directions
Understanding linkage equilibrium extends beyond theoretical interest. Its implications are vast—impacting conservation biology, personalized medicine, agriculture, and infectious disease control. For instance, in population management, promoting conditions that favor LE can preserve genetic variation, bolstering resilience. Conversely, recognizing LD patterns helps identify recent selective pressures, invaluable in identifying drug resistance mutations in pathogens or beneficial alleles in crops.
Emerging Technologies and Challenges
As genome editing tools like CRISPR become more precise, understanding linkage patterns allows for targeted modifications without unintended consequences. However, challenges remain, including accurately measuring LD in highly structured or admixed populations and interpreting LD signals amid complex evolutionary histories. Continued integration of high-throughput sequencing, bioinformatics, and functional genomics promises to unlock deeper understanding of linkage dynamics, further enhancing efforts in evolutionary conservation and medical genetics.
| Relevant Category | Substantive Data |
|---|---|
| Gene Editing | Knowledge of linkage patterns guides the selection of safe edit sites and minimizes off-target effects |
| Population Management | Maintaining or disrupting linkage equilibria influences genetic diversity conservation strategies |
| Medical Genomics | LD maps inform association studies and the development of personalized therapies |
Why is linkage equilibrium important for genetic diversity?
+Linkage equilibrium facilitates the independent assortment of alleles, which increases genetic variability and enhances adaptive potential in populations. It allows beneficial combinations to form through recombination, supporting evolution and resilience.
How does linkage disequilibrium affect evolution?
+LD can signify recent selection, genetic drift, or population structure. It can slow the shuffling of alleles, reduce genetic variation, and indicate adaptive events or population bottlenecks.
What factors influence the transition to linkage equilibrium?
+Recombination rate, population size, migration, mutation, and selection all influence how quickly a population reaches linkage equilibrium, with higher recombination and larger population sizes generally accelerating this process.